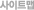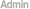H > 서비스 > Next-Generation Sequencing (NGS) > RNA-Seq(Transcriptome)
RNA-Seq(Transcriptome)
Prokaryotes RNA-Seq (Transcriptome)
Prokayotes RNA-Seq(Transcriptome)은 실험 대상 미생물체내에서 발현되는 모든 transcript (mRNA, non-coding RNA 포함)에 대한 발현 정량화 (quantification) 및 구조 분석 (strucutural analysis)을 결과물로 얻을 수 있는 NGS 기법입니다.
Prokaryotes RNA-Seq(Transcriptome) 방법을 이용하여, 해당 미생물체내의 transcriptome에 대한 대부분의 정보를 얻으실 수 있으며, 이를 이용하여 기초 분야 뿐 아니라 의학 및 약물 유전체학 등 다양한 분야에 적용될 수 있습니다.
적용분야
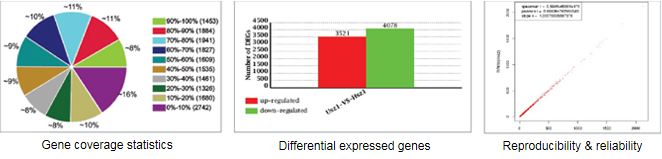
- Transcriptome 시퀀싱 (>95% coverage)
- novel coding / non-coding RNA 발굴
- gene structural analysis (alternative splicing, fusion genes)
- transcriptome 내의 SNP 분석
- differential gene expression analysis
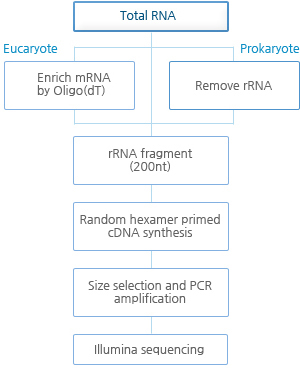
실험 Workflow
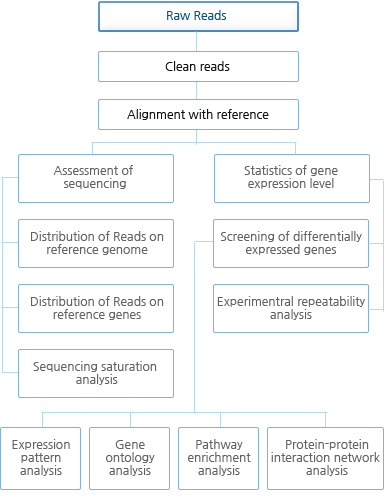
Bioinformatics 분석 Workflow
RNA-Seq(Transcriptome)-실험 진행 과정
Sample requirements
| concentration | Concentration (ng/㎕) | Quality | |
|---|---|---|---|
| Total RNA | > 2㎍ (human, mouse, rat) > 5㎍ (other animals) |
> 80 (human, mouse, rat) > 200 (other animals) |
OD(260/230) > 2.0, OD(260/280)>1.8 is highly recommended |
Sequencing Strategy
| 50 SE sequencing |
Bioinformatic analysis - contents
| RNA-Seq(Transcriptome)-resequencing | |
|---|---|
| 1차 분석 | 1. Data filtering includes removing adaptors, contamination and low-quality reads from raw reads
2. Assessment of sequencing(Statistics of raw reads, Sequencing saturation analysis, Analysis of the distribution of reads on reference gene) 3. Gene expression annotation |
| 2차 분석 | 4. Differential gene expression analysis (Screening of differentially expressed genes(DEGs), Experimental repeatability analysis of DEGs. 5. Expression pattern analysis of DEGs (Client to determine what conditions to meet to perform cluster analysis, for example, "hormone treatment in 2 hours, 4 hours, 8 hours showed a significant difference of gene expression") 6. Gene ontology analysis of DEGs 7. Pathway enrichment analysis of DEGs 8. Protein-protein interaction network analysis (protein-protein interaction database of the target species needed) |
| 3차 분석 | 별도 논의 |