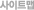H > 서비스 > Next-Generation Sequencing (NGS) >Bisulfite-Seq
Bisulfite-Seq
Bisulfite-Seq ?
Epigenetics의 한 분야인 methylation 현상을 연구하기 위해 수 많은 방법이 개발되어 왔습니다. Bisufite treatment를 이용한 방법은 가장 안정적이고 실험 재현성이 뛰어난 기법으로 알려져 있습니다.
그러므로, Bisulfite 처리한 genomic DNA에 대한 whole genome resequencing은 현재까지 개발된 methylation 연구 방법 중 가장 정확하고 재현성/반복성이 뛰어나며, 분석된 데이터는 실험 대상 생체내의 methylation 정도를 가장 잘 반영하고 있습니다.
적용분야
- Genome-wide methylation study
- differential methylated region (DMR) 탐색
- gene expression data와 연계한 통합 분석
- cancer biomarker discovery
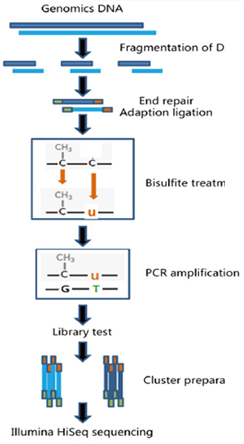
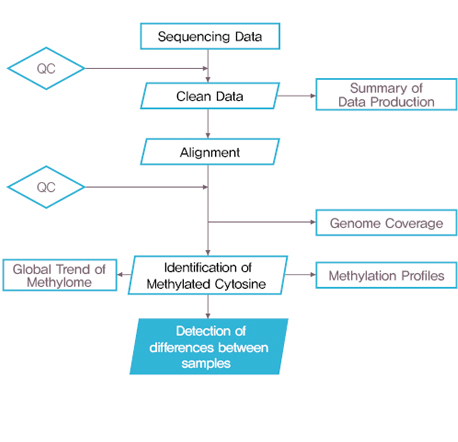
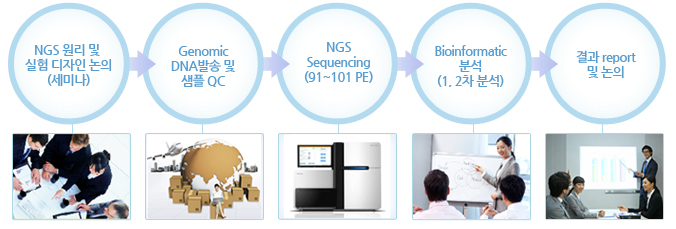
실험 Workflow
Bioinformatics 분석 Workflow
Bisulfite-Seq - 실험 진행 과정
Sample requirements
| concentration | Concentration (ng/㎕) | Quality | |
|---|---|---|---|
| Genomic DNA | > 15㎍ (in general) | > 50 | OD(260/280)>1.8 is highly recommended |
Sequencing Strategy
| 91 PE (paired ends) sequencing (50 SE or 101 SE/PE sequencing (if needed)) |
Bioinformatic analysis - contents
| contents | |
|---|---|
| 1차 분석 |
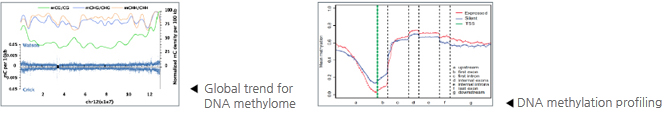
1. Data Filtering 2. Reads Alignment 3. Sequence Depth and Coverage Analysis 3.1 Cumulative Distribution of Effective Sequencing Depth in Cytosine 3.2 The Relationship between Genome Coverage and Reads Depth 4. Calculation of Methylation Level 4.1 Average Methylation Level for C, CG, CHG and CHH 4.2 Effective Coverage Analysis in Cytosine 5. Global Trends of Methylome 5.1 Proportion of Total Methyl-cytosines, CG, CHG and CHH 5.2 Methylation Level Distribution for CG, CHG and CHH 5.3 Methylation Level of Cytosines (CG, CHG, CHH) for Chromosomes 5.4 Methylation Level of Cytosines in Different Genome Regions 5.5 Methylation Level of Cytosines in Different Genomic Elements 5.6 Non-CG Proximal Sequence Features Analysis 6. Genome-wide Methylation Profiling 6.1 Methyl-cytosine Density of Each Chromosome (only for species with complete chromosomes information) 6.2 Methyl-cytosine Density of Each Scaffold (only for species with incomplete chromosomes information) 6.3 Methylation Heatmap in Different Genomic Feautures 6.4 DNA Methylation Patterns Across The Transcriptional Units at Whole Genome Level 7. Identification of Differentially Methylated Regions (DMRs) (Based on 1 and 2) |