H > 서비스 > Next-Generation Sequencing (NGS) > 16s rDNA tagging
16s rDNA tagging
16s rDNA tagging ?
특정 환경 및 시료 (stool, body fluid) 내에 존재하는 세균총의 동정, 분류 및 기능유전체 분석에 사용되는 기법입니다. 시료내의 미생물의 모든 genomic DNA를 분리하여 미생물의 종 특이적 부위로 알려져 있는 16s rDNA의 hypervariable region을 sequencing을 통하여 data를 얻은 후 미생물 동정/분류에 활용하는 방법입니다.16s rDNA는 약 1,500 bp의 길이에 해당되고, whole genome metagenomic survey에 비해 정확도가 비교적 낮지만, 저렴한 비용으로 전체 세균총의 분류를 수행할 수 있는 것이 장점입니다.
적용분야
- Metagenomics
- Microorganism classification
- Microorganism population study
- Species identification
Metagenomic survey vs. 16s rDNA tagging
| Sequencing target |
Sequencing methods |
Data | |
|---|---|---|---|
| 16s rRNA tagging |
16s rRNA hypervariable regions |
Roche 454 | 1.Taxonomic classification
2.OTU analysis
|
| Whole genome metagenomic survey |
Whole bacterial genomes |
Illumina HiSeq | 1.Species identification
2.Gene prediction
3.ORF/functional annotation |

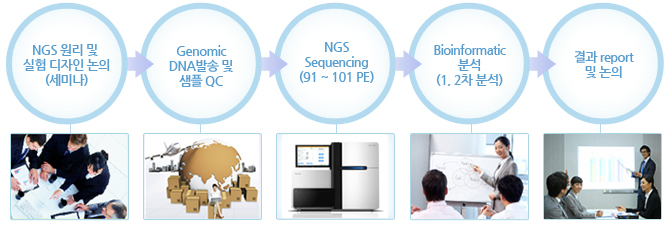
16s rDNA tagging - 실험 진행 과정
Sample requirements
| concentration | Concentration (ng/㎕) | Quality | |
|---|---|---|---|
| Genomic DNA | > 5㎍ (in general) | > 50 | OD(260/280)>1.8 is highly recommended |
Sequencing Strategy
| 91 ~ 101 PE sequencing |
Bioinformatic analysis - contents
| contents | |
|---|---|
| 1차 분석 |
1. Illumina QC report 2. Remove the low-quality reads (poly-ATGC) and adaptors 3. Overlap the paired-end reads to form tags 4. Remove the primer |
| 2차 분석 | 1. Species classification analysis by BlastN, OTU analysis 2. Calculate alpha diversity for complexity analysis in single sample 3. Species (or OTU) ralative abundance analysis 4. Primary comparative analysis: 5. Intermediate comparative analysis : 6. Advanced comparative analysis (Sample Groups ≥ 2; Samples in each Group ≥ 10): |