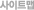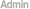H > 서비스 > Next-Generation Sequencing (NGS) > Small RNA
Small RNA
Small RNA sequencing ?
Micro RNA는 21~23 nt의 길이를 가지는 non-coding RNA이고 유전자 발현의 조절 기능을 하는 epigenetic factor의 하나로 알려져 있습니다. Small RNA sequencing을 통하여, 현재까지 알려진 micro RNA의 발현 변화 연구뿐 아니라, 알려지지 않은 새로운 micro RNA를 발굴할 수 있습니다. 또한, sequencing 결과와 생명 현상과의 연계성 및 치료제 개발 등에도 적용될 수 있습니다.
적용분야
- Small RNA expression analysis
- Small RNA differential expression analysis
- novel small RNA discovery
- Application to biomarkers and drug targets
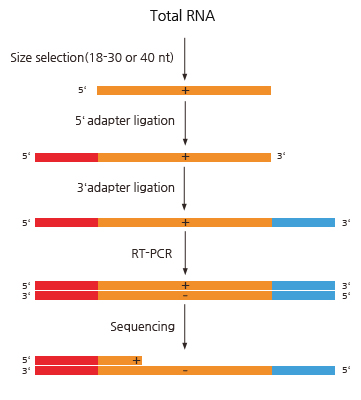
실험 Workflow
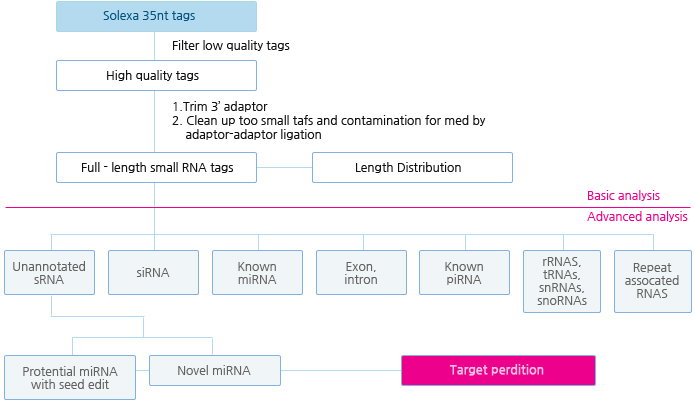
Bioinformatics 분석 Workflow
Small RNA sequencing - 실험 진행 과정
Sample requirements
| concentration | Concentration (ng/㎕) | Quality | |
|---|---|---|---|
| Total RNA | > 10㎍ (human, mouse, rat) |
> 80 | OD(260/230) > 2.0, OD(260/280)>1.8 is highly recommended |
| Small RNA | > 1㎍ (mirVana miRNA isolation kit)> | > 20 |
Sequencing Strategy
| 50 SE sequencing |
Bioinformatic analysis - contents
| contents | |
|---|---|
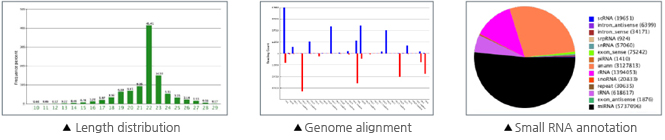
| 1차 분석 | 1. Remove adaptors, low quality tags as well as contaminants to get clean reads 2. Summarize the length distribution of small RNA 3. Analyze common and specific sequences between two samples 4. Explore small RNA distribution across selected genome 5. Identify rRNAs, tRNAs, snRNAs, etc. by aligning to Rfam and Genbank databases 6. Identify repeat associated small RNAs 7. Identify degradated fragments of mRNAs 8. Identify known miRNAs by aligning to designated part of miRBase 9. Annotate small RNAs into several categories based on priority 10. Predict novel miRNAs and their secondary structures by Mireap from unannotated small RNAs 11. Analyze the expression pattern of known miRNAs 12. Family analysis of known miRNAs |
| 2차 분석 | 13. Differential expression analysis of miRNA and cluster analysis (three or more samples should be provided) 14. Target genes prediction of miRNA(the sequence of coding genes should be provided) 15. Target genes GO annotation and KEGG pathway analysis of miRNA 16. Base editing analysis of miRNA(known miRNA) |
| 3차 분석 | 별도 논의 |