H > 서비스 > Next-Generation Sequencing (NGS) > ChIP-Seq
ChIP-Seq
ChIP-Seq ?
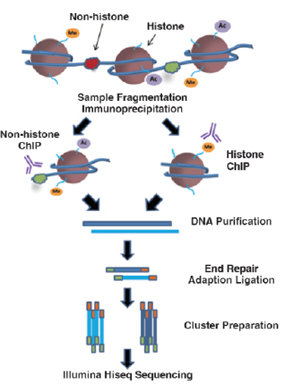
특정 DNA binding motif (transcription factor 등)의 DNA binding site를 whole genome level에서 스크리닝하기 위한 방법으로, 해당 motif의 antibody를 이용하여 ChIP (chromatin IP)를 수행한 후, enrichment된 DNA fragment를 sequencing하여 binding site를 연구하는 방법입니다. 주로 transcription factor와 histone modification 연구에 적용되고 있습니다.
적용분야
- Transcription factor binding site 연구
- Histone modification 연구
- Protein-DNA interaction 연구
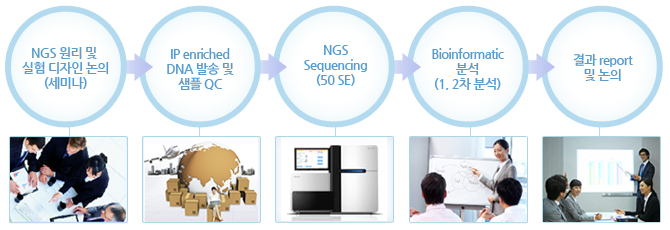
실험 Workflow
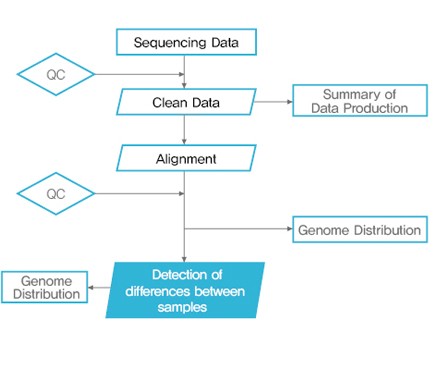
Bioinformatics 분석 Workflow
ChIP-Seq - 실험 진행 과정
Sample requirements
| concentration | Concentration (ng/㎕) | Quality | |
|---|---|---|---|
| IP enriched DNA | > 10㎍ (in general) | > 1 | OD(260/280)>1.8 is highly recommended . DNA fragment : between 100 bp ~ 500bp |
Sequencing Strategy
| 50 PE (paired ends) sequencing (50 SE, 91 PE and 101 SE/PE sequencing (if needed)) |
Bioinformatic analysis - contents
| contents | |
|---|---|
| 1차 분석 |
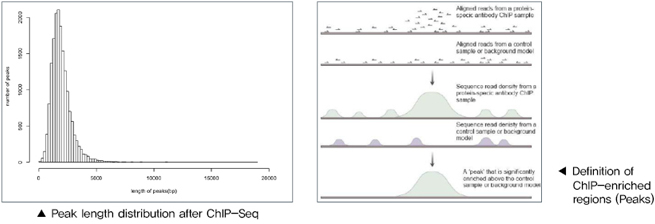
1. Data Filtering 2. Reads Alignment 3. Genome-wide Distribution of ChIP Sequencing Reads 4. Peak Scanning and Distribution 5. Peak-related Genes Scanning and GO Function Analysis 6. Difference Analysis of Multi-samples 7. UCSC Genome Browser instruction |
| 2차 분석 | 1. ChIP Sequencing Reads Distribution around TSS 2. Motif analysis 2.1 Candidate Motif Scanning 2.2 Known Motif Analysis (database should be provided by customer) |